Click here to get instructions…
- Please download and unzip the replication files for Chapter 6 ( Chapter06.zip).
- Read
readme.htmland run6-0_ChapterSetup.R. This will create6-0_ChapterSetup.RDatain the sub folderdata/R. This file contains the data required to produce the table shown at the bottom of this page. - We also recommend to load the libraries listed in the Chapter 6’s
LoadInstallPackages.R
# assuming you are working within .Rproj environment
library(here)
# install (if necessary) and load other required packages
source(here("source", "LoadInstallPackages.R"))
# load environment generated in "6-0_ChapterSetup.R"
load(here("data", "R", "6-0_ChapterSetup.RData"))
Share of explained discrepancy
Table 6.2 in Chapter 6.2.1 shows how much of the
dicrepancy in the labor market (activity.year.seq) and
family (partner.child.year.seq) sequences is explained by
the three dummy variables east (living in East vs West
Germany), sex (male vs. female), highschool
(at least highschool degree: yes vs. no). The dummy variables are stored
in the data frames family and activity.
When considering only one variable at a time, {TraMineR}’s dissassoc
function is the fastest solution for computing the share of explained
discrepancy (pseudo-\(R^2\)). With
dissmfacw {TraMineR} also provides the
possibility to consider multiple variables at a time.
Table 6.2 includes both a univariable and a mutlivariable analysis of the three variables. As the grouping variables do not explain the discrepancy of the family biographies the multivariable approach is only presented for the labor market sequences.
| \(R^2\) | p-value | \(R^2\) | p-value | \(\Delta R^2\) | p-value | |
|---|---|---|---|---|---|---|
| Region | 0.01 | 0.001 | 0.01 | 0.001 | 0.00 | 0.001 |
| Gender | 0.01 | 0.001 | 0.09 | 0.001 | 0.09 | 0.001 |
| Highschool degree | 0.01 | 0.001 | 0.07 | 0.001 | 0.06 | 0.001 |
| \(R^2_{total}\) | ||||||
| Total | 0.16 | 0.001 |
Like in Chapter 6.1 we wrote a function and several lines of code to
extract the results from the {TraMineR} functions and to print a
nice table using {knitr}’s kable and the
{kableExtra} package. You can find the code in the script
6-2_Table_6-2_R2.R stored in
Chapter06.zip).
On this page, however, we do not further elaborate on such
technicalities and rather focus on briefly showcasing the plain {TraMineR} commands required to obtain
the results shown in the table. As you see, this does not require a lot
of coding:
# Labor market sequences
dissassoc(activity.year.om, activity$sex)
dissassoc(activity.year.om, activity$east)
dissassoc(activity.year.om, activity$highschool)
dissmfacw(activity.year.om ~ east + sex + highschool,
data = activity)
# Family sequences
dissassoc(partner.child.year.om, family$sex)
dissassoc(partner.child.year.om, family$east)
dissassoc(partner.child.year.om, family$highschool)Extracting specifically the information needed for the table requires
some additional commands. The following code chunks briefly illustrate
the general procedure. We first save and inspect the results from a
discrepancy analysis of activity.year.om by gender
(activity$sex).
discr_sex <- dissassoc(activity.year.om, activity$sex)
discr_sexPseudo ANOVA table:
SS df MSE
Exp 981.5653 1 981.565295
Res 10027.9985 1025 9.783413
Total 11009.5638 1026 10.730569
Test values (p-values based on 1000 permutation):
t0 p.value
Pseudo F 100.32953529 0.001
Pseudo Fbf 101.24714023 0.001
Pseudo R2 0.08915569 0.001
Bartlett 31.59488891 0.001
Levene 172.45072755 0.001
Inconclusive intervals:
0.00383 < 0.01 < 0.0162
0.03649 < 0.05 < 0.0635
Discrepancy per level:
n discrepancy
0 506 7.346404
1 521 12.112702
Total 1027 10.720121{TraMineR}’s dissassoc
produces a lot of output. We save the output in the object
discr_sex. A closer inspection of this object with
str or names shows that dissassoc
stores its results in a list object with seven elements. The information
required for Table 6.2 - pseudo-\(R^2\) and the corresponding p-value - is
stored in the list element stat.
str(discr_sex)List of 7
$ groups :'data.frame': 3 obs. of 2 variables:
..$ n : num [1:3] 506 521 1027
..$ discrepancy: num [1:3] 7.35 12.11 10.72
$ anova.table :'data.frame': 3 obs. of 3 variables:
..$ SS : num [1:3] 982 10028 11010
..$ df : num [1:3] 1 1025 1026
..$ MSE: num [1:3] 981.57 9.78 10.73
$ perms :List of 4
..$ R : num 1000
..$ t0 : num [1:5] 100.3295 101.2471 0.0892 31.5949 172.4507
..$ t : num [1:1000, 1:5] 100.33 0.688 1.222 0.589 0.828 ...
..$ pval: num [1:5] 0.001 0.001 0.001 0.001 0.001
..- attr(*, "class")= chr "TraMineRPermut"
$ stat :'data.frame': 5 obs. of 2 variables:
..$ t0 : num [1:5] 100.3295 101.2471 0.0892 31.5949 172.4507
..$ p.value: num [1:5] 0.001 0.001 0.001 0.001 0.001
$ weight.permutation: chr "none"
$ call : language dissassocweighted(diss = diss, group = group, weights = weights, R = R, weight.permutation = weight.permutat| __truncated__
$ R : num 1000
- attr(*, "class")= chr "dissassoc"names(discr_sex) [1] "groups" "anova.table" "perms"
[4] "stat" "weight.permutation" "call"
[7] "R" discr_sex$stat t0 p.value
Pseudo F 100.32953529 0.001
Pseudo Fbf 101.24714023 0.001
Pseudo R2 0.08915569 0.001
Bartlett 31.59488891 0.001
Levene 172.45072755 0.001If we want to extract pseudo-\(R^2\), for instance, we can type:
discr_sex$stat[3,1] %>%
round(2)[1] 0.09The script 6-2_Table_6-2_R2.R illustrates all the steps
required to extract, arrange, and format the output from
dissassoc and dissmfacw as shown in
Table 6.2.
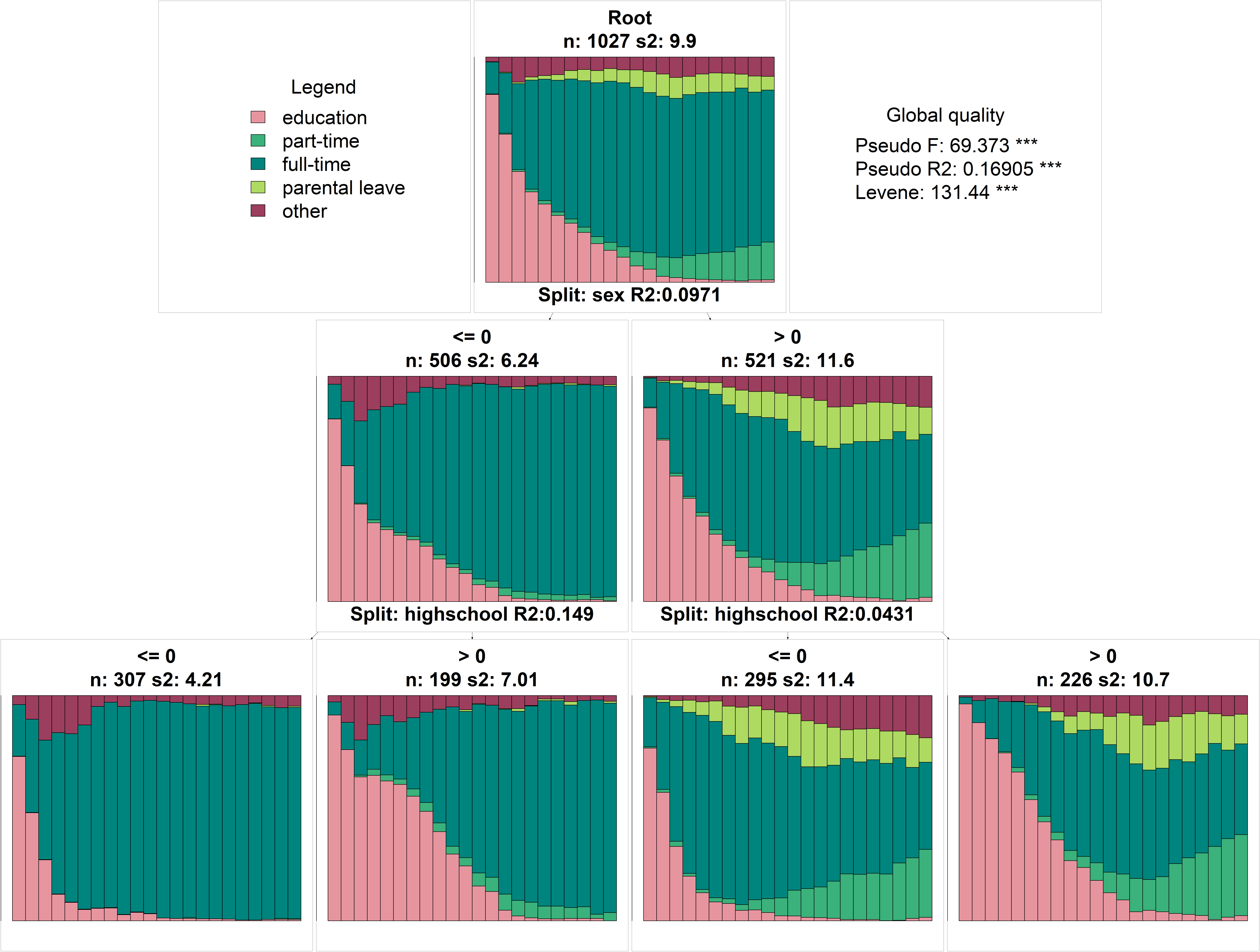
Regression Tree
The regression tree displayed in Figure 6.1 is based
on an analysis of labor market sequences with a reduced alphabet
distinguishing 5 instead of 8 states. We define the new sequence object
by recoding the original sequence object activity.year.seq
stored in 6-0_ChapterSetup.RData with {TraMineR}’s seqrecode
function. Note that you have to take care of the labels after you
defined a new sequence object with seqrecode.
# Inspect the original alphabet
alphabet(activity.year.seq)[1] "EDU" "MIL/CS" "PT" "FT" "SELF" "PLEAVE"
[7] "MARGINAL" "UNEMP" # Recode alphabet
activity.year.seq2 <- seqrecode(activity.year.seq,
recodes = list("EDU" = "EDU",
"PT" = "PT",
"FT" = c("FT", "SELF"),
"PLEAVE" = "PLEAVE",
"OTHER" = c("MIL/CS","MARGINAL", "UNEMP")))
# Specify labels for new alphabet
attributes(activity.year.seq2)$labels <- c("education",
"part-time", "full-time",
"parental leave", "other")The regression tree shown in Figure 6.1 is based on
a regression tree analysis done with {TraMineR}’s seqtree. The
function requires to specify a regression like formula
(activity.year.seq2 ~ east + sex + highschool) and a
dissimilarity matrix (activity.year.om2) telling it which
variables should be considered for partitioning a given sequence object.
The function offers several opportunities to set cut-off criteria
restricting the further growth of the tree. In the example below we
restrict the depth of the tree to three (max.depth = 3),
i.e. to branches with a maximum of two splits of the starting partition
(the original sequence object).
# Compute dissimilarity matrix required as input for regression tree
activity.year.om2 <- seqdist(activity.year.seq2,
method="OM", sm= "CONSTANT")
# Run regression tree analysis
activitytree <- seqtree(activity.year.seq2 ~ east + sex + highschool,
data = activity, diss = activity.year.om2,
weighted = F, max.depth = 3)
# Print the tree
activitytreeDissimilarity tree:
Parameters: min.size=51.35, max.depth=3, R=1000, pval=0.01
Formula: activity.year.seq2 ~ east + sex + highschool
Global R2: 0.16905
Fitted tree:
|-- Root (n: 1027 disc: 9.9024)
|-> sex 0.097106
|-- <= 0 (n: 506 disc: 6.2448)
|-> highschool 0.14948
|-- <= 0 (n: 307 disc: 4.2088)[(EDU,2)-(FT,20)] *
|-- > 0 (n: 199 disc: 7.0122)[(EDU,7)-(FT,15)] *
|-- > 0 (n: 521 disc: 11.559)
|-> highschool 0.043057
|-- <= 0 (n: 295 disc: 11.353)[(EDU,2)-(FT,16)-(PLEAVE,3)-(PT,1)] *
|-- > 0 (n: 226 disc: 10.681)[(EDU,8)-(FT,10)-(PLEAVE,1)-(PT,3)] * Although, the printed tree provides a lot of information a graphical
visualization of the tree is more appealing and insightful. The
visualization can be rendered with the function
seqtreedisplay and the open source graph visualization
software GraphViz.
Once you have downloaded and installed GraphViz it can be used by
seqtreedisplay. It might be necessary, however, to inform
the function where to find GraphViz on your computer
(gvpath-argument of seqtreedisplay). In the
following code chunk we first define the GraphViz path and
then render the graph.
# specify GraphViz directory
graphviz.dir <- "C:/Program Files/GraphViz"
# Plot the results and save the figure
seqtreedisplay(activitytree, type = "d",
cex.main = 2.5,
with.legend=T,
gvpath = graphviz.dir,
filename = here("figures", "Figure_6-1_Tree.png"))